A
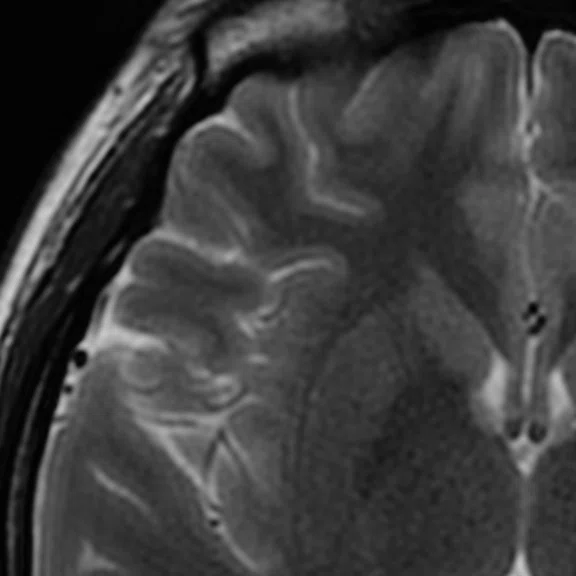
Figure 1.
Axial T2 FSE, 1 NEX, in 50 sec. (A) without AIR™ Recon DL and (B) with AIR™ Recon DL.
B
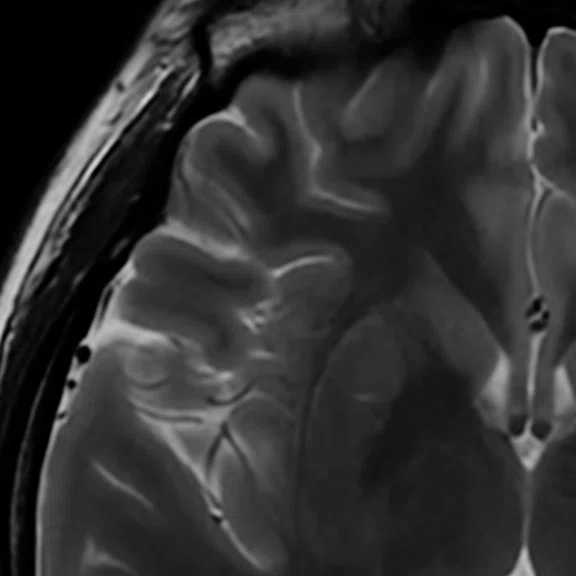
Figure 1.
Axial T2 FSE, 1 NEX, in 50 sec. (A) without AIR™ Recon DL and (B) with AIR™ Recon DL.
‡ Technology in development that represents ongoing research and development efforts. These technologies are not products and may never become products. Not for sale. Not cleared or approved by the US FDA or any other global regulator for commercial availability.
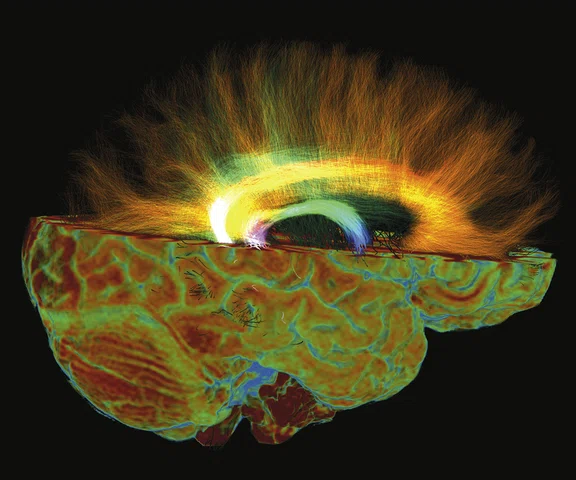
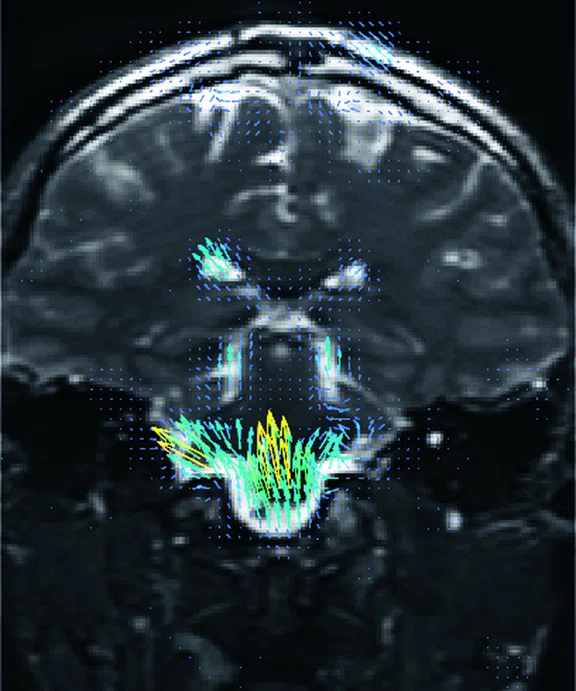
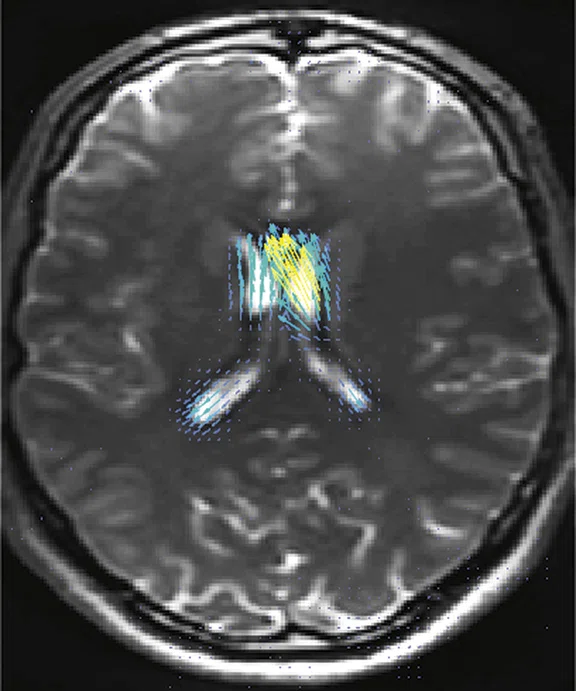
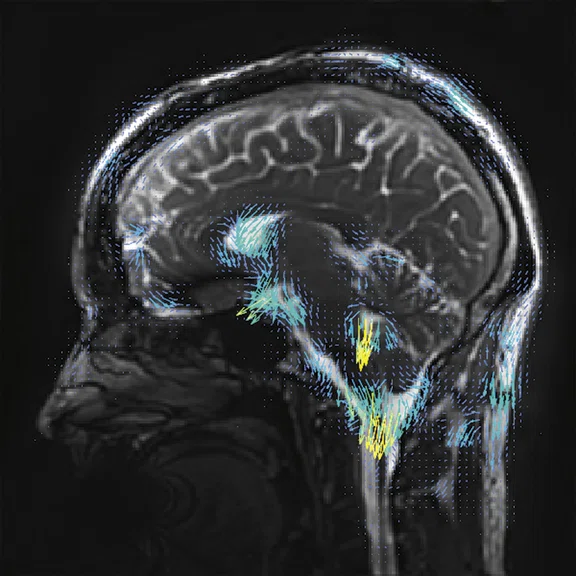
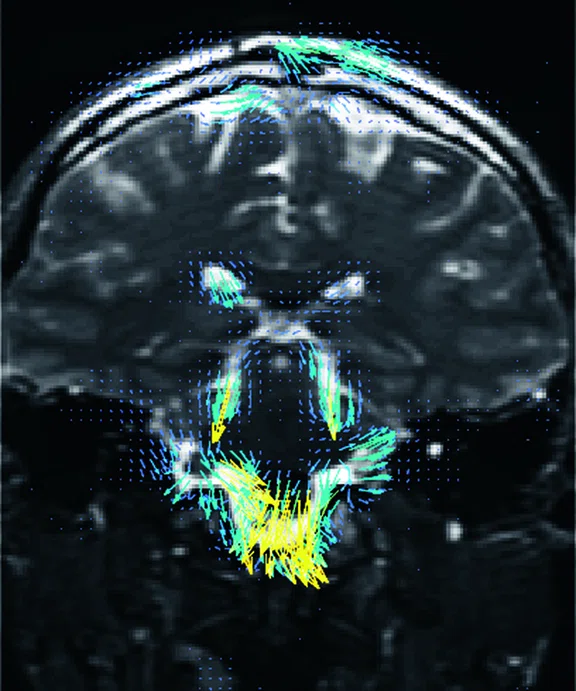
Figure 2.
Higher order tensor diffusion MR acquired on a SIGNA™ Premier 3.0T system. Data
were acquired with a diffusion sequence set up by Dr. Jerome Maller with parameters: 2 mm3, with 3 b-values = 1000, 2000, 3000 s/mm2, 15, 15, 30 directions respectively, 4 b=0, scan time = 3:40 min.). Images are processed using High Angular Resolution Diffusion Imaging (HARDI) Probabilistic Tractography, with 100,000 streamlines for each bundle of tracts. MRTrix was used for the tractography. Images post-processed by Maryam Tayebi from Auckland Bioengineering Institute and Mātai.
1. Holdsworth SJ, Rahimi MS, Ni WW, Zaharchuk G, Moseley ME. Amplified magnetic resonance imaging (aMRI). Magn Reson Med. 2016 Jun;75(6):2245-54. doi: 10.1002/mrm.26142.
2. Terem I, Ni WW, Goubran M, et al. Revealing sub-voxel motions of brain tissue using phase-based amplified MRI (aMRI). Magn Reson Med. 2018 Dec;80(6):2549-2559. doi: 10.1002/mrm.27236.
3. Terem I, Dang L, Champagne A, et al. 3D amplified MRI (aMRI). Magn Reson Med. 2021 Sep;86(3):1674-1686. doi: 10.1002/mrm.28797.
4. Abderezaei J, Pionteck A, Terem I, et al. Development, calibration, and validation of 3D aMRI for the quantification of intrinsic brain motion. Brain Multiphysics, (2021). 100022, ISSN 2666-5220. doi: 10.1016/j.brain.2021.100022.
3. Terem I, Dang L, Champagne A, et al. 3D amplified MRI (aMRI). Magn Reson Med. 2021 Sep;86(3):1674-1686. doi: 10.1002/mrm.28797.
4. Abderezaei J, Pionteck A, Terem I, et al. Development, calibration, and validation of 3D aMRI for the quantification of intrinsic brain motion. Brain Multiphysics, (2021). 100022, ISSN 2666-5220. doi: 10.1016/j.brain.2021.100022.
A
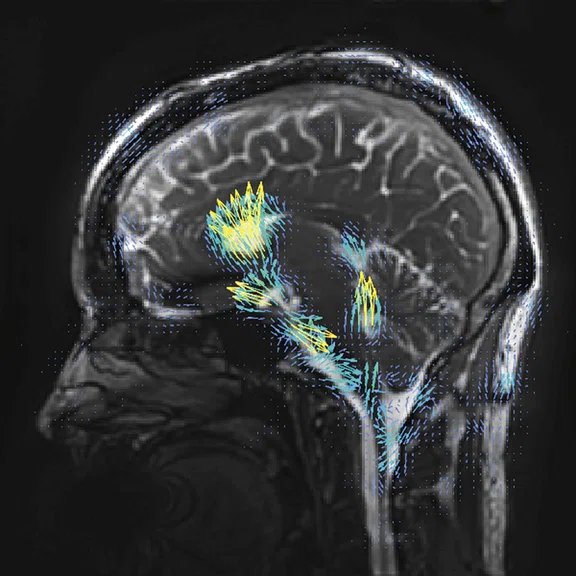
Figure 3.
3D aMRI acquired on a SIGNA™ Premier 3.0T system. Data were acquired with GE’s 3D bSSFP Cine sequence, processed with the 3D aMRI algorithm to exaggerate brain motion. The amplified data have been processed by optical flow (small arrows) to show general tissue motion. The whole brain 1 mm isotropic dataset was acquired in a scan time of 1:40 min. (A-C) Cardiac frame 13 and (D-F) cardiac frame 17.
B
Figure 3.
3D aMRI acquired on a SIGNA™ Premier 3.0T system. Data were acquired with GE’s 3D bSSFP Cine sequence, processed with the 3D aMRI algorithm to exaggerate brain motion. The amplified data have been processed by optical flow (small arrows) to show general tissue motion. The whole brain 1 mm isotropic dataset was acquired in a scan time of 1:40 min. (A-C) Cardiac frame 13 and (D-F) cardiac frame 17.
C
Figure 3.
3D aMRI acquired on a SIGNA™ Premier 3.0T system. Data were acquired with GE’s 3D bSSFP Cine sequence, processed with the 3D aMRI algorithm to exaggerate brain motion. The amplified data have been processed by optical flow (small arrows) to show general tissue motion. The whole brain 1 mm isotropic dataset was acquired in a scan time of 1:40 min. (A-C) Cardiac frame 13 and (D-F) cardiac frame 17.
D
Figure 3.
3D aMRI acquired on a SIGNA™ Premier 3.0T system. Data were acquired with GE’s 3D bSSFP Cine sequence, processed with the 3D aMRI algorithm to exaggerate brain motion. The amplified data have been processed by optical flow (small arrows) to show general tissue motion. The whole brain 1 mm isotropic dataset was acquired in a scan time of 1:40 min. (A-C) Cardiac frame 13 and (D-F) cardiac frame 17.
E
Figure 3.
3D aMRI acquired on a SIGNA™ Premier 3.0T system. Data were acquired with GE’s 3D bSSFP Cine sequence, processed with the 3D aMRI algorithm to exaggerate brain motion. The amplified data have been processed by optical flow (small arrows) to show general tissue motion. The whole brain 1 mm isotropic dataset was acquired in a scan time of 1:40 min. (A-C) Cardiac frame 13 and (D-F) cardiac frame 17.
F
Figure 3.
3D aMRI acquired on a SIGNA™ Premier 3.0T system. Data were acquired with GE’s 3D bSSFP Cine sequence, processed with the 3D aMRI algorithm to exaggerate brain motion. The amplified data have been processed by optical flow (small arrows) to show general tissue motion. The whole brain 1 mm isotropic dataset was acquired in a scan time of 1:40 min. (A-C) Cardiac frame 13 and (D-F) cardiac frame 17.
result


PREVIOUS
${prev-page}
NEXT
${next-page}
Subscribe Now
Manage Subscription
FOLLOW US
Contact Us • Cookie Preferences • Privacy Policy • California Privacy PolicyDo Not Sell or Share My Personal Information • Terms & Conditions • Security
© 2024 GE HealthCare. GE is a trademark of General Electric Company. Used under trademark license.
SPOTLIGHT
Finding connections: New Zealand institute using advanced MR protocol in search of mTBI imaging biomarkers
Finding connections: New Zealand institute using advanced MR protocol in search of mTBI imaging biomarkers
In the rural town of Gisborne, New Zealand, is a world-class MR research center working to develop a reliable method for diagnosing the presence and severity of mild traumatic brain injury (mTBI), often referred to as a concussion.
In the rural town of Gisborne, New Zealand, is a world-class MR research center working to develop a reliable method for diagnosing the presence and severity of mild traumatic brain injury (mTBI), often referred to as a concussion.
Samantha Holdsworth, PhD, returned to her hometown from Stanford University and founded the Mātai Medical Research Institute, a not-for-profit research center focused on enhancing the capabilities of medical imaging using new and advanced software, post-processing and artificial intelligence.
"All the work we do at Mātai is designed to make an impact in this community and beyond," says Dr. Holdsworth. "We can make an immediate difference and provide advanced medical imaging to people who typically don’t have that type of access."
While Dr. Holdsworth and her colleagues may be geographically isolated, they are connected to technical and clinical experts throughout the world through their partnership with GE Healthcare.
"It turns out, if you partner with the right people you are just one phone call away to reach scientists, developers and application specialists all over the world," says Daniel Cornfeld, MD, clinical lead at Mātai. "That’s the sort of infrastructure you normally need a large university to have. So, we’ve been very fortunate to have GE and the University of Auckland in our corner."
Drs. Holdsworth and Cornfeld knew they wanted the strongest, fastest gradients on a 3.0T MR system that also offered advanced coils and sequences. During their search, they learned about AIR™ 48-channel Head Coil, AIR™ Recon DL and HyperBand from GE. Seeing these solutions in action at GE MR corporate headquarters and the impact of these three core technologies on reducing scan times without sacrificing SNR and image quality solidified their decision to select the SIGNA™ Premier, GE’s leading, innovative 3.0T MR scanner for advanced imaging and research.
"When we realized how fast we could go imaging the brain, heart and prostate, that opened up the potential for a lot of projects," Dr. Cornfeld adds. "Having an advanced scanner with great image quality that could go fast and help with throughput was important."
The research
Mātai’s flagship research study is investigating imaging biomarkers for mTBI. There are currently no reliable ways to diagnose the presence or severity of mTBI.
As part of the study, Mātai is evaluating a cohort of secondary school rugby players pre-season, mid-season, post-season and after any suspected in-season concussion. All subjects wear mouthguards with accelerometers and undergo a clinical evaluation, saliva testing, acceleration analysis and eye tracking; but the centerpiece of the study is a one hour, comprehensive, MR exam on SIGNA™ Premier.
“This is a very advanced MR protocol for examining the brain for signs of mTBI. We have at least eight or nine sequences that we can capture in about one hour because we can go so fast, and that is a real accomplishment.”
Dr. Samantha Holdsworth
It is expected that other mTBI research centers will adapt these study protocols developed at Mātai. The protocol includes high-resolution 3D T1 BRAVO and 3D Cube FLAIR, parameter mapping with MAGiC, 3D arterial spin labeling (ASL), resting-state and task-based functional MRI (fMRI), quantitative susceptibility mapping (QSM)‡, multishell diffusion, time of flight (TOF) angiography, multi-voxel spectroscopy of the corpus callosum, 4D-phase contrast and a newly developed technique called amplified MRI (aMRI)‡.
The rugby players are outfitted with kinematic mouth guard sensors to track their head impacts, enabling the researchers to create models of brain injury under impact and in near real time.
"Diffusion MR is one of our most important sequences because it already shows quite a bit of promise in picking up a brain injury due to concussion," Dr. Holdsworth says. While the SIGNA™ Premier is equipped with a multi-shell diffusion MR protocol, Drs. Holdsworth and Cornfeld wanted to push the limits in acquiring data as quickly as possible.
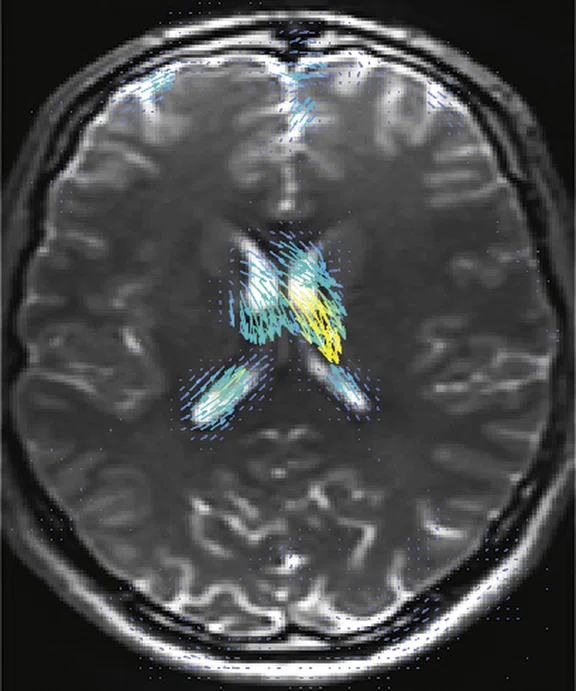
For image post-processing support, they turned to Jerome Maller, PhD, GE MR Clinical Science Specialist, who developed a multi-shell, three b-value DTI research protocol‡ with interleaved b-zero volumes to generate high spatial and angular resolution data in a very short time. The DTI sequence used as part of the study runs 50 directions at 2 mm resolution with 3D values, all in under 4 minutes (Figure 2).
Figure 2.
Higher order tensor diffusion MR acquired on a SIGNA™ Premier 3.0T system. Data
were acquired with a diffusion sequence set up by Dr. Jerome Maller with parameters: 2 mm3, with 3 b-values = 1000, 2000, 3000 s/mm2, 15, 15, 30 directions respectively, 4 b=0, scan time = 3:40 min.). Images are processed using High Angular Resolution Diffusion Imaging (HARDI) Probabilistic Tractography, with 100,000 streamlines for each bundle of tracts. MRTrix was used for the tractography. Images post-processed by Maryam Tayebi from Auckland Bioengineering Institute and Mātai.
"It is just spectacular," Dr. Holdsworth adds. "It is difficult to resolve kissing fiber tracks that are hard to see with one b-value. We believe that it will give us higher accuracy in our fractional anisotropy and tractography—the images coming out of these data sets are just absolutely stunning."
Capturing all the DTI data this fast enables the researchers to add in other promising sequences, such as QSM, 3D ASL, multi-voxel spectroscopy, fMRI and anatomical sequences, as well.
Specialized modifications were also made to the SWAN sequence, utilizing different control variables to make it a QSM sequence, following some of the work being done by Yi Wang, PhD, at Cornell University.
Another important sequence is aMRI, which enables the researchers to observe the biomechanical response of the brain by utilizing a motion magnification algorithm with a cine MR acquisition1-4. Dr. Holdsworth’s team, comprised of a group of international collaborators, are applying their latest 3D aMRI algorithm to amplified cardiac gated balanced steady-state free precession (bSSFP)‡ acquisitions to improve the image quality and visualization of cardiac and cerebral spinal fluid (CSF) induced brain motion in three directions3,4.
With the 3D bSSFP Cine sequence accelerated by a factor of 8, Mātai can acquire a whole brain acquisition with a 1 × 1 × 1 mm resolution in under 2 minutes and use this to amplify the brain motion in 3D with exquisite resolution (Figure 3).
Figure 3.
3D aMRI acquired on a SIGNA™ Premier 3.0T system. Data were acquired with GE’s 3D bSSFP Cine sequence, processed with the 3D aMRI algorithm to exaggerate brain motion. The amplified data have been processed by optical flow (small arrows) to show general tissue motion. The whole brain 1 mm isotropic dataset was acquired in a scan time of 1:40 min. (A-C) Cardiac frame 13 and (D-F) cardiac frame 17.
"Previously at Stanford, the scan time would have been considerably longer on the older software platform – we’ve been really impressed by how fast we can go with this sequence now," says Dr. Holdsworth. With this sequence, the researchers are looking for subtle changes in the brain mechanics, such as a difference in the motion of the brain due to blood flow, CSF, etc., and in mTBI which may create a change in brain pressure due to brain swelling.
"Because it is such a short sequence, we run that sequence on everyone to see the variety of brain motion across populations so we can provide control data for applications such as idiopathic intracranial hypertension, TBI and other applications where there may be a change in brain pressure and therefore motion and tissue viability," Dr. Holdsworth says.
Mātai and GE are working together to help share the aMRI post-processing algorithm to research sites throughout the world.
Dr. Cornfeld is also interested in examining whether a 3D version‡ of MAGiC can replace some of the anatomical imaging being conducted on subjects in the study. Currently, the anatomic sequences take up to 7 minutes. If Mātai can synthetically create the 3D axial T1 BRAVO and 3D sagittal FLAIR data sets with 3D MAGiC, then they can save up to 5 minutes; time that can be used to add another functional sequence.
"That’s an advantage of working closely with GE, because as advancements in MR sequences become available, we can update the protocol so it continues to get better," adds Dr. Cornfeld.
Mātai is currently in the post-season scanning stage, completing the data collection on approximately 66% of the study participants. Athletes who participate in other non-contact sports, such as swimming and basketball, are also being scanned with the same advanced MR imaging protocol as healthy controls. Next, the team will analyze the data and report their findings. However, one thing is already clear to Dr. Holdsworth.
"We should be able to do a much better job of estimating the brain impact from a hit to the head with the better quality imaging data we are obtaining on the SIGNA™ Premier," she says. "One of the issues with concussion is that it’s very hard to diagnose the presence or absence of it because it manifests different clinical symptoms. There is a real need for an objective biomarker of concussion, whether that be axon or chemical damage, increased lactate or something else."
Adds Dr. Cornfeld, "If we want to stratify severity of the concussion, we first need a really good way of diagnosing the injury and then study the effects of what that injury is. Having a proper biomarker will allow us and others to do all that research."